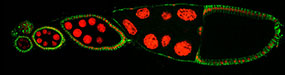
We have studied the molecular mechanisms responsible for setting up the body plan of the Drosophila embryo. This patterning process relies on deployment of several key proteins at specific locations in the oocyte and embryo. Deployment of the proteins is acheived by a combination of mechanisms:
- mRNA localization
- repression and activation of the translation of the localized mRNAs
- anchoring of the proteins encoded by the localized mRNAs
Projects in the lab have addressed these mechanisms, primarily localization of bicoid mRNA, and localization and translational regulation of oskar mRNA.
During this work we have often focused on the cis-acting sequences present in mRNAs that direct their localization and translational status, and the proteins that bind these sequences. We identify, map and characterize the cis-acting elements to learn about their roles. Knowing that a certain cis-acting element performs an important and interesting function, we then look for the protein that binds the element to mediate this function. Through genetic and biochemical characterization of such a binding protein we can determine if it does indeed mediate the function of the cis-acting element, and begin to understand how it acts.